Bayesian optimization
Suppose we want to optimize a function such as validation error (which is a
function of hyperparameters) but it’s really complicated. We can approximate
it with a simple function, called the surrogate function. After we have queried a certain number of points, we can condition on
these point to infer the posterior over the surrogate function using Bayesian
linear regression.
Bayesian Optimization iteratively uses a probabilistic surrogate model and an acquisition function to determine next point of evaluation in a search space. At each iteration step, the surrogate model is fit to all observations of the target function. The acquisition function determines the utility of the different candidate points using the predictive distribution of the probabilistic model. Bayesian optimization has the advantage of been derivative-free optimizer which means in situations where the function to be optimized has a derivative that is intractable or is not continuous, it can still be optimized by this method. It is also high performing,in fact experiments published in this paper here by Turner et al. shows that several implementations of Bayesian Optimization for the purpose of hyperparameter tuning outperforms the random search and grid search methods.
Bayesian hyperparameter optimization is a trade off between exploration (hyperparameters for which the outcome is most uncertain) and exploitation (hyperparameters expected close to the optimum). Some AutoML algorithms employ bayesian optimization for automatic hyperparameter tuning.
In Bayesian optimization, the surrogate function that quantifies the uncertainty in the objective function is optimized in place of the actual objective function. This is done when the objective function is costly to optimize. Bayesian optimization allows for global minimization of black-box functions without evaluating the derivative of the function. The bayesian approach treats the objective function as a random function and places a prior distribution on it based on our knowledge of the function. An acquisition function defined from this surrogate is used to decide where to sample.
There are several packages for bayesian optimization in both Python and R. In this work we will implement several of such libraries applied to the the problem of hyperparameter tuning of machine learning models. Two models the XGBoost and Lightgbm algorithms will be considered. These two are among the best performing and most popular Tree-based gradient boosting ensemble models around.
Common Libraries for Bayesian Hyperparameter Tuning.
There are many frameworks you can use to bayesian optimization in Python for bayesian-optimization, these include bayesian-optimization, Hyperopt, Scikit-Optimize, Optuna etc. The focus of this work will be to show how you can perform bayesian optimization in applied in the context of hyperparameter tuningn of XGBoost and LightGBM using the frameworks listed below. The idea is to model the hyperparameter search process probabilistically. An evaluation metric AUC or F1 value is used select the next combination of hyper-parameters, such that the improvement in the metric is maximum. We would illustrate how to perform bayesian optimization applied in the context hyperparameter tuning using the following libraries. The libraries below are the most common and easy to implement bayesian optimizers. The goal is to have a comprehensive resources that can get one started quickly on tuning hyperparameters using bayesian optimization.
- Scikit-optimize
- Hyperopt
- bayesian-optimization
- optuna
Data
https://archive.ics.uci.edu/ml/datasets/census+income
1. Using The BayesianOptimization Package
First install the libraries required for the model building ,eveluation and hyperparameter tuning.
#!pip install kaggledatasets
#import kaggledatasets as kd
#!pip install -q kaggle
#dataset = kd.CreditCardFraudDetection(download=True)
#dataset.head()
cols=\
['age','workclass','fnlwgt','education','education-num','marital-status','occupation','relationship','race','sex','capital-gain','capital-loss','hours-per-week','native-country','Income']
%%capture
# Install helper functions.
!pip install catboost -U
!pip install talos
!pip install bayesian-optimization==1.0.1
!pip install -q git+https://github.com/gmihaila/ml_things.git
!pip install skll
!pip install imblearn
!pip install hyperopt
There is some incompatibility issues between latest versions scikit-learn and scikit-optimize. These older versions work well together.
!pip install -Iv scikit-learn==0.22.1
!pip install scikit-optimize==0.7.4
Requirement already satisfied: scikit-optimize==0.7.4 in /usr/local/lib/python3.7/dist-packages (0.7.4)
Requirement already satisfied: scikit-learn>=0.19.1 in /usr/local/lib/python3.7/dist-packages (from scikit-optimize==0.7.4) (0.23.2)
Requirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.7/dist-packages (from scikit-optimize==0.7.4) (1.0.1)
Requirement already satisfied: numpy>=1.11.0 in /usr/local/lib/python3.7/dist-packages (from scikit-optimize==0.7.4) (1.21.1)
Requirement already satisfied: scipy>=0.18.0 in /usr/local/lib/python3.7/dist-packages (from scikit-optimize==0.7.4) (1.7.0)
Requirement already satisfied: pyaml>=16.9 in /usr/local/lib/python3.7/dist-packages (from scikit-optimize==0.7.4) (20.4.0)
Requirement already satisfied: PyYAML in /usr/local/lib/python3.7/dist-packages (from pyaml>=16.9->scikit-optimize==0.7.4) (3.13)
Requirement already satisfied: threadpoolctl>=2.0.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn>=0.19.1->scikit-optimize==0.7.4) (2.2.0)
import xgboost as xgb
from bayes_opt import BayesianOptimization
from sklearn.metrics import mean_squared_error
import pandas as pd
import numpy as np
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from xgboost import XGBClassifier
from sklearn.metrics import classification_report, confusion_matrix
% autosave 5
from google.colab import drive
drive.mount('/drive')
Mounted at /content/drive
import pandas as pd
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from xgboost import XGBClassifier
#Make directory named kaggle and copy kaggle.json file there.
train_data = pd.read_csv("/Data/adult.data.zip",header=None,names=cols)
train_data.head()
| age | workclass | fnlwgt | education | education-num | marital-status | occupation | relationship | race | sex | capital-gain | capital-loss | hours-per-week | native-country | Income | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 39 | State-gov | 77516 | Bachelors | 13 | Never-married | Adm-clerical | Not-in-family | White | Male | 2174 | 0 | 40 | United-States | <=50K |
| 1 | 50 | Self-emp-not-inc | 83311 | Bachelors | 13 | Married-civ-spouse | Exec-managerial | Husband | White | Male | 0 | 0 | 13 | United-States | <=50K |
| 2 | 38 | Private | 215646 | HS-grad | 9 | Divorced | Handlers-cleaners | Not-in-family | White | Male | 0 | 0 | 40 | United-States | <=50K |
| 3 | 53 | Private | 234721 | 11th | 7 | Married-civ-spouse | Handlers-cleaners | Husband | Black | Male | 0 | 0 | 40 | United-States | <=50K |
| 4 | 28 | Private | 338409 | Bachelors | 13 | Married-civ-spouse | Prof-specialty | Wife | Black | Female | 0 | 0 | 40 | Cuba | <=50K |
test_data = pd.read_csv("/Data/adult.test",skiprows=1,header=None,names=cols)
test_data.head()
| age | workclass | fnlwgt | education | education-num | marital-status | occupation | relationship | race | sex | capital-gain | capital-loss | hours-per-week | native-country | Income | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 25 | Private | 226802 | 11th | 7 | Never-married | Machine-op-inspct | Own-child | Black | Male | 0 | 0 | 40 | United-States | <=50K. |
| 1 | 38 | Private | 89814 | HS-grad | 9 | Married-civ-spouse | Farming-fishing | Husband | White | Male | 0 | 0 | 50 | United-States | <=50K. |
| 2 | 28 | Local-gov | 336951 | Assoc-acdm | 12 | Married-civ-spouse | Protective-serv | Husband | White | Male | 0 | 0 | 40 | United-States | >50K. |
| 3 | 44 | Private | 160323 | Some-college | 10 | Married-civ-spouse | Machine-op-inspct | Husband | Black | Male | 7688 | 0 | 40 | United-States | >50K. |
| 4 | 18 | ? | 103497 | Some-college | 10 | Never-married | ? | Own-child | White | Female | 0 | 0 | 30 | United-States | <=50K. |
categorical_columns = train_data.select_dtypes(include=['object']).columns
le = LabelEncoder()
#y= le.fit_transform(df.label)
train_data[categorical_columns] = train_data[categorical_columns].apply(le.fit_transform)
test_data[categorical_columns] = test_data[categorical_columns].apply(le.fit_transform)
train_data.head()
| age | workclass | fnlwgt | education | education-num | marital-status | occupation | relationship | race | sex | capital-gain | capital-loss | hours-per-week | native-country | Income | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 39 | 7 | 77516 | 9 | 13 | 4 | 1 | 1 | 4 | 1 | 2174 | 0 | 40 | 39 | 0 |
| 1 | 50 | 6 | 83311 | 9 | 13 | 2 | 4 | 0 | 4 | 1 | 0 | 0 | 13 | 39 | 0 |
| 2 | 38 | 4 | 215646 | 11 | 9 | 0 | 6 | 1 | 4 | 1 | 0 | 0 | 40 | 39 | 0 |
| 3 | 53 | 4 | 234721 | 1 | 7 | 2 | 6 | 0 | 2 | 1 | 0 | 0 | 40 | 39 | 0 |
| 4 | 28 | 4 | 338409 | 9 | 13 | 2 | 10 | 5 | 2 | 0 | 0 | 0 | 40 | 5 | 0 |
Training BayesianOptimization
import xgboost as xgb
from bayes_opt import BayesianOptimization
from sklearn.metrics import mean_squared_error
import pandas as pd
import numpy as np
from xgboost import XGBClassifier
Create XGBoost dataset here
from sklearn.model_selection import train_test_split
#X_train, X_test, y_train, y_test = train_test_split(df.drop('fare_amount', axis=1),df['Income'], test_size=0.25)
#del(df)
#Converting the dataframe into XGBoost’s Dmatrix object
dtrain = xgb.DMatrix(train_data.drop('Income', axis=1),label=train_data.Income)
#del(X_train)
dtest = xgb.DMatrix(test_data.drop('Income', axis=1),label=test_data.Income)
#del(X_test)
Formulate the machine learning hyperparameter tunining as an optimization problem with an objective function evaluate. Here the parameters to be tuned are max_depth, gamma and colsample_bytree. A three fold cross validation is used used in the hyperparameter tuning process. The return value of this function is the mean of the cross-validation auc.
For regression problems, return of this function is the mean squared error. It is multiplied by negative one because the optimization process will by default be a maximization process but we need the minimum mean squared error as the best model or in other words the multiplication by -1 converts the optimization maximization problem to minimization problem
import gc
import warnings
from bayes_opt import BayesianOptimization
from sklearn.model_selection import cross_val_score, StratifiedKFold, StratifiedShuffleSplit
from sklearn.metrics import log_loss, matthews_corrcoef, roc_auc_score
from sklearn.preprocessing import MinMaxScaler
import xgboost as xgb
import contextlib
#The cross-va;lidation parameters is saved into the log file
log_file = open('xgb_opt.log', 'a')
# Comment out any parameter you don't want to test
def xgb_evaluate(
max_depth,
gamma,
min_child_weight,
max_delta_step,
subsample,
colsample_bytree,
num_boost_round
):
#
# Define all XGboost parameters
#
#
params = {
'booster' : 'gbtree',
'max_depth' : int(max_depth),
'gamma' : gamma,
'eta' : 0.1,
'objective' : 'binary:logistic',
'num_boost_round':num_boost_round,
'nthread' : -1,
'silent' : True,
'eval_metric': 'auc',
'subsample' : max(min(subsample, 1), 0),
'colsample_bytree' : max(min(colsample_bytree, 1), 0),
'min_child_weight' : min_child_weight,
'max_delta_step' : int(max_delta_step),
'seed' : 1001
}
folds = 5
cv_score = 0
print("\n Search parameters (%d-fold validation):\n %s" % (folds, params), file=log_file )
log_file.flush()
#Cross validating with the specified parameters in 5 folds and 70 iterations
cv_result = xgb.cv(
params,
dtrain,
#num_boost_round = 70,
stratified = True,
nfold = folds,
# verbose_eval = 10,
early_stopping_rounds = 10,
metrics = 'auc',
show_stdv = True
)
val_score = cv_result['test-auc-mean'].iloc[-1]
train_score = cv_result['train-auc-mean'].iloc[-1]
print(' Stopped after %d iterations with train-auc = %f val-auc = %f ( diff = %f ) train-gini = %f val-gini = %f' % ( len(cv_result), train_score, val_score, (train_score - val_score), (train_score*2-1),
(val_score*2-1)) )
# Bayesian optimization only knows how to maximize, not minimize, so return the negative RMSE
#in case you are tuning a regression model
#return -1.0 * cv_result['test-rmse-mean'].iloc[-1]
return (val_score*2) - 1
Here we specify the domain values for the search space of the parameters we are tuning. Use the expected improvement acquisition function to handle negative numbers.Optimally needs quite a few more initiation points and number of iterations.
xgb_bo = BayesianOptimization(xgb_evaluate, {
'max_depth': (2, 12),
'gamma': (0.001, 10.0),
'min_child_weight': (0, 20),
'max_delta_step': (0, 10),
'subsample': (0.4, 1.0),
'num_boost_round':(20,100),
'colsample_bytree' :(0.2, 1.0)
})
print('-'*130)
print('-'*130, file=log_file)
#The flush() method cleans out the internal buffer.
log_file.flush()
with warnings.catch_warnings():
warnings.filterwarnings('ignore')
#performing Bayesian optimization for 5 iterations with 8 steps of random
#exploration with an #acquisition function of expected improvement
xgb_bo.maximize(init_points=2, n_iter=5, acq='ei', xi=0.0)
----------------------------------------------------------------------------------------------------------------------------------
| iter | target | colsam... | gamma | max_de... | max_depth | min_ch... | num_bo... | subsample |
-------------------------------------------------------------------------------------------------------------
Stopped after 10 iterations with train-auc = 0.914569 val-auc = 0.910411 ( diff = 0.004158 ) train-gini = 0.829138 val-gini = 0.820822
| [0m 1 [0m | [0m 0.8208 [0m | [0m 0.7703 [0m | [0m 3.676 [0m | [0m 6.028 [0m | [0m 6.732 [0m | [0m 2.524 [0m | [0m 71.26 [0m | [0m 0.9426 [0m |
Stopped after 10 iterations with train-auc = 0.910387 val-auc = 0.902667 ( diff = 0.007720 ) train-gini = 0.820774 val-gini = 0.805334
| [0m 2 [0m | [0m 0.8053 [0m | [0m 0.4517 [0m | [0m 2.845 [0m | [0m 1.253 [0m | [0m 11.57 [0m | [0m 14.76 [0m | [0m 77.97 [0m | [0m 0.8765 [0m |
Stopped after 10 iterations with train-auc = 0.865323 val-auc = 0.865657 ( diff = -0.000333 ) train-gini = 0.730647 val-gini = 0.731314
| [0m 3 [0m | [0m 0.7313 [0m | [0m 0.2 [0m | [0m 10.0 [0m | [0m 10.0 [0m | [0m 2.0 [0m | [0m 0.0 [0m | [0m 100.0 [0m | [0m 0.4 [0m |
Stopped after 10 iterations with train-auc = 0.867601 val-auc = 0.867338 ( diff = 0.000263 ) train-gini = 0.735202 val-gini = 0.734676
| [0m 4 [0m | [0m 0.7347 [0m | [0m 1.0 [0m | [0m 0.001 [0m | [0m 10.0 [0m | [0m 2.0 [0m | [0m 20.0 [0m | [0m 70.0 [0m | [0m 1.0 [0m |
Stopped after 10 iterations with train-auc = 0.900196 val-auc = 0.895841 ( diff = 0.004355 ) train-gini = 0.800392 val-gini = 0.791682
| [0m 5 [0m | [0m 0.7917 [0m | [0m 0.2 [0m | [0m 10.0 [0m | [0m 0.0 [0m | [0m 12.0 [0m | [0m 0.0 [0m | [0m 70.0 [0m | [0m 1.0 [0m |
Stopped after 10 iterations with train-auc = 0.941382 val-auc = 0.911827 ( diff = 0.029556 ) train-gini = 0.882765 val-gini = 0.823653
| [95m 6 [0m | [95m 0.8237 [0m | [95m 1.0 [0m | [95m 0.001 [0m | [95m 0.0 [0m | [95m 12.0 [0m | [95m 0.0 [0m | [95m 83.26 [0m | [95m 0.4 [0m |
Stopped after 10 iterations with train-auc = 0.911175 val-auc = 0.895202 ( diff = 0.015972 ) train-gini = 0.822350 val-gini = 0.790405
| [0m 7 [0m | [0m 0.7904 [0m | [0m 0.2 [0m | [0m 0.001 [0m | [0m 10.0 [0m | [0m 12.0 [0m | [0m 0.0 [0m | [0m 70.0 [0m | [0m 1.0 [0m |
=============================================================================================================
results = pd.DataFrame(xgb_bo.res)
f"maximum Gini {max(results.target)}"
print('-'*130)
print('Final Results')
best_params = results[results.target == max(pd.DataFrame(xgb_bo.res).target)]['params']
print('best_params {}'.format(best_params.values))
#log_file.flush()
#log_file.close()
f"best AUC {( max(results.target) + 1 ) / 2}"
----------------------------------------------------------------------------------------------------------------------------------
Final Results
best_params [{'colsample_bytree': 1.0, 'gamma': 0.001, 'max_delta_step': 0.0, 'max_depth': 12.0, 'min_child_weight': 0.0, 'num_boost_round': 83.25554188122533, 'subsample': 0.4}]
'best AUC 0.9118266'
Testing
After the hyperparameter tuning, we can train a final model with the best hyperparameters from the search.
#Extracting the best parameters
#type(best_params.values)
params = dict(best_params)
print(params)
param = {'colsample_bytree': 1.0,
'gamma': 0.001,
'max_delta_step': 0.0,
'max_depth': 12,
'objective': 'binary:logistic',
'min_child_weight': 0.0,
#'num_boost_round': 83,
'n_estimators': 83,
'subsample': 0.4}
#Converting the max_depth and n_estimator values from float to int
#params['max_depth']= int(params['max_depth'])
#params['n_estimators']= int(params['n_estimators'])
#Initialize an XGBClassifier with the tuned parameters and fit the training data
from xgboost import XGBClassifier
classifier = XGBClassifier( max_depth=12, learning_rate=0.1, n_estimators=83,
silent=True, objective='binary:logistic',
booster='gbtree')
#final_model = classifier.fit(train_data.drop('Income', axis=1),train_data.Income)
classifier2 = XGBClassifier(**param).fit(train_data.drop('Income', axis=1),train_data.Income)
#predicting for training set
pred = classifier2.predict(test_data.drop('Income', axis=1))
#Looking at the classification report
print(classification_report(pred, test_data.Income))
{5: {'colsample_bytree': 1.0, 'gamma': 0.001, 'max_delta_step': 0.0, 'max_depth': 12.0, 'min_child_weight': 0.0, 'num_boost_round': 83.25554188122533, 'subsample': 0.4}}
precision recall f1-score support
0 0.94 0.89 0.91 13220
1 0.61 0.77 0.68 3061
accuracy 0.87 16281
macro avg 0.78 0.83 0.80 16281
weighted avg 0.88 0.87 0.87 16281
#Attained prediction accuracy on the training set
cm = confusion_matrix(pred, test_data.Income)
acc = cm.diagonal().sum()/cm.sum()
print(acc)
0.865794484368282
# Predict on holdout set
test = transform(test)
dtest = xgb.DMatrix(test)
y_pred_test = model2.predict(dtest)
Bayesian Hyperparameter Tuning of Lightgbm Model
We repeat the hyperparameter tuning process above but this time with a LightGBM model.The lightgbm is another well known tree-based boosting algorithm. LightGBM is a gradient boosting framework that uses tree based learning algorithms. It’s advantages include lower memory usgae, faster training speed, better accuracy and support of parallel or distributed training.
#train_data = lgb.Dataset(data, label=label, weight=w)
import lightgbm as lgb
lgbdtrain = lgb.Dataset(train_data.drop('Income', axis=1), label=train_data.Income)
folds =5
def lgb_evaluate(max_depth, colsample_bytree,scale_pos_weight,num_boost_round):
params = {'metric': 'auc',
'max_depth': int(max_depth),
'subsample': 0.8,
'objective' : 'binary',
'learning_rate': 0.1,
# ' importance_type'='gain',
'colsample_bytree': colsample_bytree,
'scale_pos_weight' : scale_pos_weight,
'num_boost_round': int(num_boost_round)}
# Used around 1000 boosting rounds in the full model
cv_result = lgb.cv(params,
lgbdtrain,
#num_boost_round=70, #same as n_estimators
stratified = True,
nfold = folds,
# verbose_eval = 10,
early_stopping_rounds = 10,
metrics = 'auc',
show_stdv = True
)
# Bayesian optimization only knows how to maximize, not minimize, so return the negative RMSE
#when building a regression model.
return cv_result['auc-mean'][-1]
lgb_bo = BayesianOptimization(lgb_evaluate, {'max_depth': (3, 10),
#'gamma': (0, 1),
'colsample_bytree': (0.3, 0.9),
'scale_pos_weight' : (1,100),
'num_boost_round': (50,150)},
verbose=5 # verbose = 1 prints only when a maximum is observed, verbose = 0 is silent
)
# Use the expected improvement acquisition function to handle negative numbers
# Optimally needs quite a few more initiation points and number of iterations
lgb_bo.maximize(init_points=10, n_iter=5, acq='ei')
| iter | target | colsam... | max_depth | num_bo... | scale_... |
-------------------------------------------------------------------------
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 1 [0m | [0m 0.9263 [0m | [0m 0.5399 [0m | [0m 8.415 [0m | [0m 120.4 [0m | [0m 56.75 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 2 [0m | [0m 0.9246 [0m | [0m 0.6526 [0m | [0m 9.683 [0m | [0m 86.06 [0m | [0m 71.91 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [95m 3 [0m | [95m 0.9285 [0m | [95m 0.6556 [0m | [95m 9.617 [0m | [95m 124.9 [0m | [95m 6.988 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 4 [0m | [0m 0.9199 [0m | [0m 0.727 [0m | [0m 5.297 [0m | [0m 63.09 [0m | [0m 92.59 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 5 [0m | [0m 0.9244 [0m | [0m 0.629 [0m | [0m 8.541 [0m | [0m 67.81 [0m | [0m 49.62 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 6 [0m | [0m 0.9219 [0m | [0m 0.7857 [0m | [0m 5.621 [0m | [0m 67.87 [0m | [0m 82.2 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 7 [0m | [0m 0.9253 [0m | [0m 0.6557 [0m | [0m 5.685 [0m | [0m 83.85 [0m | [0m 38.17 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 8 [0m | [0m 0.9252 [0m | [0m 0.7254 [0m | [0m 8.538 [0m | [0m 148.4 [0m | [0m 99.16 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 9 [0m | [0m 0.9252 [0m | [0m 0.5192 [0m | [0m 9.303 [0m | [0m 108.6 [0m | [0m 94.68 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 10 [0m | [0m 0.9246 [0m | [0m 0.5287 [0m | [0m 7.356 [0m | [0m 80.54 [0m | [0m 59.86 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [95m 11 [0m | [95m 0.929 [0m | [95m 0.739 [0m | [95m 9.401 [0m | [95m 149.3 [0m | [95m 1.108 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [95m 12 [0m | [95m 0.9293 [0m | [95m 0.4673 [0m | [95m 9.957 [0m | [95m 149.7 [0m | [95m 1.431 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 13 [0m | [0m 0.9288 [0m | [0m 0.7267 [0m | [0m 9.851 [0m | [0m 149.9 [0m | [0m 1.301 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 14 [0m | [0m 0.9289 [0m | [0m 0.6839 [0m | [0m 9.349 [0m | [0m 150.0 [0m | [0m 1.035 [0m |
/usr/local/lib/python3.7/dist-packages/lightgbm/engine.py:430: UserWarning: Found `num_boost_round` in params. Will use it instead of argument
warnings.warn("Found `{}` in params. Will use it instead of argument".format(alias))
| [0m 15 [0m | [0m 0.9292 [0m | [0m 0.3656 [0m | [0m 9.803 [0m | [0m 149.2 [0m | [0m 1.432 [0m |
=========================================================================
results = pd.DataFrame(lgb_bo.res)
f"maximum AUC {max(results.target)}"
print('-'*130)
print('Final Results')
best_params = results[results.target == max(pd.DataFrame(lgb_bo.res).target)]['params']
print('best_params {}'.format(best_params.values))
----------------------------------------------------------------------------------------------------------------------------------
Final Results
best_params [{'colsample_bytree': 0.46727594393341354, 'max_depth': 9.957368160190926, 'num_boost_round': 149.73209643654872, 'scale_pos_weight': 1.4314916482964704}]
### 2. Using the scikit-optimize Library Here we would perform the above hyperparameter tuning with the bayes-optimize package. The bayes optimize package integrates very nicely with the scikit-learn package. This makes hyperparameter tuning of scikit-learn models a breeze. You would only need to replace GridSearchCV or RandomizedSearchCV used in scikit-learn hyperparameter tuning with BayesSearchCV class from the scikit-optimize library.
import pandas as pd
import numpy as np
from tqdm import tqdm
from scipy import stats
from scipy.stats import chi2_contingency
import sys
import pandas as pd
import io
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns; sns.set(style="ticks", color_codes=True)
from sklearn import preprocessing
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_selection import SelectKBest, f_classif
from sklearn.feature_selection import chi2
from sklearn.linear_model import LogisticRegression
from sklearn import feature_selection
from sklearn.impute import SimpleImputer
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import Pipeline
from sklearn.metrics import make_scorer
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import accuracy_score
from sklearn.model_selection import cross_validate
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_classification
from sklearn.model_selection import StratifiedKFold
from sklearn.feature_selection import RFECV
from xgboost import XGBClassifier
from sklearn.impute import SimpleImputer
import xgboost as xgb
import lightgbm as lgb
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import svm
from skopt.space import Real, Categorical, Integer
#from skll.metrics import spearman
from skopt import BayesSearchCV
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import Pipeline
from sklearn.metrics import make_scorer
from skopt.space import Real, Categorical, Integer
from sklearn.metrics import classification_report
from sklearn.base import TransformerMixin
from sklearn.metrics import classification_report, f1_score, accuracy_score, precision_score, confusion_matrix
from sklearn.metrics import roc_auc_score
from sklearn.metrics import precision_recall_curve
pd.set_option('display.max_rows', 600)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
import warnings
import pandas_profiling
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.feature_selection import SelectFromModel
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.pipeline import make_pipeline, FeatureUnion, Pipeline
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split, RandomizedSearchCV
from sklearn.metrics import roc_curve, roc_auc_score
from sklearn.svm import SVC
import random
import matplotlib.pyplot as plt
import seaborn as sns
import re
from sklearn.preprocessing import StandardScaler, OneHotEncoder
#from imblearn.over_sampling import RandomOverSampler
#from imblearn.under_sampling import RandomUnderSampler
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import *
from sklearn.utils import resample
#from imblearn.over_sampling import SMOTE
#import smote
import os
from sklearn.tree import DecisionTreeClassifier
#from imblearn.metrics import geometric_mean_score as gmean
#from imblearn.metrics import make_index_balanced_accuracy as iba
#from imblearn.metrics import *
#import rus
# Skopt functions
from skopt import BayesSearchCV
from skopt import gp_minimize # Bayesian optimization using Gaussian Processes
from skopt.space import Real, Categorical, Integer
from skopt.utils import use_named_args # decorator to convert a list of parameters to named arguments
from skopt.callbacks import DeadlineStopper # Stop the optimization before running out of a fixed budget of time.
from skopt.callbacks import VerboseCallback # Callback to control the verbosity
from skopt.callbacks import DeltaXStopper # Stop the optimization If the last two positions at which the objective has been evaluated are less than delta
from joblib import dump, load
from prettytable import PrettyTable
from collections import Counter
from sklearn.datasets import make_classification
from hyperopt import fmin, tpe, space_eval,hp
from bayes_opt import BayesianOptimization
import skopt
import sklearn
import gc
warnings.filterwarnings("ignore")
%matplotlib inline
%autosave 2
print('The scikit-optimize version is {}.'.format(skopt.__version__))
print('The scikit-learn version is {}.'.format(sklearn.__version__))
imputer = SimpleImputer(missing_values=np.nan, strategy='mean')
Autosaving every 2 seconds
The scikit-optimize version is 0.7.4.
The scikit-learn version is 0.22.1.
XGBoost Scikit-optimize
We will attempt to tune an xgboost model here with the scikit-optimize library.
# Setting a 5-fold stratified cross-validation (note: shuffle=True)
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=0)
clf = xgb.XGBClassifier(
n_jobs = -1,
objective = 'binary:logistic',
silent=1,
tree_method='approx')
search_spaces = {'learning_rate': Real(0.1, 1.0, 'log-uniform'),
'min_child_weight': Integer(0, 10),
'max_depth': Integer(1,12),
'max_delta_step': Integer(0, 10), # Maximum delta step we allow each leaf output
'subsample': Real(0.5, 1.0, 'uniform'),
'colsample_bytree': Real(0.1, 1.0, 'uniform'), # subsample ratio of columns by tree
'colsample_bylevel': Real(0.1, 1.0, 'uniform'), # subsample ratio by level in trees
#'reg_lambda': Real(1e-9, 1000, 'log-uniform'), # L2 regularization
#'reg_alpha': Real(1e-9, 1.0, 'log-uniform'), # L1 regularization
'gamma': Real(1e-1, 0.5, 'log-uniform'), # Minimum loss reduction for partition
'n_estimators': Integer(50, 200),
#'scale_pos_weight': Real(1e-6, 500, 'log-uniform'), this parameter allows to build a
#cost-sensitive model to handle target class imbalance.
'scale_pos_weight': Real(1, 1000)}
bayessearch = BayesSearchCV(clf,
search_spaces,
scoring='f1',
cv=skf,
n_iter=40,
n_jobs=-1,
return_train_score=True,
refit=True,
optimizer_kwargs={'base_estimator': 'GP'},
random_state=22)
xgbm_model = bayessearch.fit(X=train_data.drop('Income', axis=1),y=train_data.Income)
print("val. score: %s" % bayessearch.best_score_)
print("test score: %s" % bayessearch.score(test_data.drop('Income', axis=1), test_data.Income))
print("best params: %s" % str(bayessearch.best_params_))
val. score: 0.7064478652278865
test score: 0.6891364096629539
best params: OrderedDict([('colsample_bylevel', 0.8074985755754681), ('colsample_bytree', 1.0), ('gamma', 0.21433018756345557), ('learning_rate', 0.1), ('max_delta_step', 10), ('max_depth', 10), ('min_child_weight', 10), ('n_estimators', 200), ('scale_pos_weight', 1.0), ('subsample', 0.9648072349277772)])
dump(xgbm_model, '/xgb_model.joblib')
xgbm_model = load('/xgb_model.joblib')
# Create the evaluation report.
evaluation_report = classification_report(true_labels, predictions_labels, labels=list(labels_ids.values()), target_names=list(labels_ids.keys()))
# Show the evaluation report.
print("Classification Report")
print(evaluation_report)
# Plot confusion matrix.
plot_confusion_matrix(y_true=true_labels, y_pred=predictions_labels,
classes=list(labels_ids.keys()), normalize=True,
magnify=1,
);
Lightgbm Hyperparameter Optimization scikit-optimize
Here we attempt to tune the lightgbm model using the scikit-optimize library
#train_data=pd.concat([cat_enc_df,num_df],axis=1)
#categorical_feat=[i for i in x_train_imp.columns.tolist() if i in catdf.columns.tolist()]
#lgb_model_ros = lgb_estimator.fit(X=X_ros, y=y_ros,feature_name=list(x_train_imp.columns),
# categorical_feature=categorical_feat)
RANDOM_STATE = 148
clf=lgb.LGBMClassifier( random_state=RANDOM_STATE,
importance_type='gain')
# the modeling pipeline
pipe = Pipeline([("imputer", SimpleImputer()),
("scaler" ,MinMaxScaler()),
("estimator", clf)])
# Setting a 5-fold stratified cross-validation (note: shuffle=True)
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=0)
search_spaces = {
'imputer__strategy': Categorical(['median']),
'estimator__learning_rate': Real(0.01, 1.0, 'log-uniform'),
'estimator__num_leaves': Integer(2, 100),
'estimator__max_depth': Integer(0, 500),
'estimator__min_child_samples': Integer(0, 200), # minimal number of data in one leaf
'estimator__max_bin': Integer(100, 1000), # max number of bins that feature values will be bucketed
'estimator__subsample': Real(0.01, 1.0, 'uniform'),
'estimator__subsample_freq': Integer(0, 10), # bagging fraction
'estimator__colsample_bytree': Real(0.01, 1.0, 'uniform'), # enabler of bagging fraction
'estimator__min_child_weight': Integer(1, 10), # minimal number of data in one leaf.
'estimator__subsample_for_bin': Integer(100, 500), # number of data that sampled for histogram bins
'estimator__reg_lambda': Real(1e-9, 1000, 'log-uniform'), # L2 regularization
'estimator__reg_alpha': Real(1e-9, 1.0, 'log-uniform'), # L1 regularization
'estimator__scale_pos_weight': Real(1e-6, 500, 'log-uniform'),
'estimator__n_estimators': Integer(100,400) ,
'estimator__scale_pos_weight':Integer(1,1000)
}
# create our search object
opt = BayesSearchCV(pipe,
search_spaces,
cv=5,
n_jobs=-1,
scoring='f1', #roc_auc, f1, f1_micro/macro,balaced_accuracy,recall
#is_unbalance ='True' , #used in binary class training
verbose=0,
error_score=-9999,
#scoring='average_precision',
random_state=RANDOM_STATE,
return_train_score=True,
n_iter=75)
lgb_model= opt.fit(X=train_data.drop('Income', axis=1),y=train_data.Income)
print("val. score: %s" % opt.best_score_)
print("test score: %s" % opt.score(test_data.drop('Income', axis=1), test_data.Income))
print("best params: %s" % str(opt.best_params_))
val. score: 0.6935562662753282
test score: 0.68503937007874
best params: OrderedDict([('estimator__colsample_bytree', 0.7363157688145477), ('estimator__learning_rate', 0.2998370128718745), ('estimator__max_bin', 596), ('estimator__max_depth', 371), ('estimator__min_child_samples', 0), ('estimator__min_child_weight', 1), ('estimator__n_estimators', 400), ('estimator__num_leaves', 57), ('estimator__reg_alpha', 0.9997837140809686), ('estimator__reg_lambda', 1000.0), ('estimator__scale_pos_weight', 1), ('estimator__subsample', 1.0), ('estimator__subsample_for_bin', 438), ('estimator__subsample_freq', 7), ('imputer__strategy', 'median')])
3. Using the Optuna library
Optuna uses a technique called Tree-Parzen estimator to select the set of hyper-parameters to be considered in the next iteration of the search, based on the history of experiments. It also has functionality for logging and visualizations of experiments and hyperparameters More details of this framework is available on the adevelopers page here.
To use optuna with neptune.ai which give you access to more resources to visualize the tuning process etc., you will need to first create an account at neptune. This would allow you create a project,get an API token and allow you to create an experiment on neptune in your notebook. The name of my project is bayeshyper and my experiment is bayes hpo. Our goal is to maximize auc which is the value of the objective function. The steps to tuning parameters with optuna in conjunction with neptune.ai are create an obective function which returns the value of the evaluation metric in this case AUC.Next create a named experiment, create a study with a study name. We want the experiment to sored in an sql-lite database so we specify the location as ‘sqlite:///bayeshpo.db’ whereas the study anme is “bayeshpo-experiment”. The NeptuneMonitor class from neptune.ai creates a callback that automatically logs all the useful information from the tuning process which can later be used in creating visualizations. study.optimize begins the optimization, the number of steps here N_TRIALS is set to 100 trials. It must be noted that Optuna can be used without neptune.ai. We show how to use Optuna with neptune.ai and by itself.
!pip install optuna
!pip install neptune-client
!pip install neptune-contrib
import lightgbm as lgb
import neptune.new as neptune
#import neptune
import neptunecontrib.monitoring.optuna as opt_utils
import optuna
import pandas as pd
from sklearn.model_selection import train_test_split
import neptunecontrib.monitoring.optuna as opt_utils #
!set NEPTUNE_API_TOKEN="eyJhcGlfYWRkcmVzcyI6Imh0dHBzOi8vYXBwLm5lcHR1bmUuYWkiLCJhcGlfdXJsIjoiaHR0cHM6Ly9hcHAubmVwdHVuZS5haSIsImFwaV9rZXkiOiIxNzcwN2E0Yy05M2NkLTRjZTUtOGQ1MC01NTE2MGUyZjE2ODkifQ=="
NEPTUNE_API_TOKEN ="eyJhcGlfYWRkcmVzcyI6Imh0dHBzOi8vYXBwLm5lcHR1bmUuYWkiLCJhcGlfdXJsIjoiaHR0cHM6Ly9hcHAubmVwdHVuZS5haSIsImFwaV9rZXkiOiIxNzcwN2E0Yy05M2NkLTRjZTUtOGQ1MC01NTE2MGUyZjE2ODkifQ=="
NUM_BOOST_ROUND = 100
EARLY_STOPPING_ROUNDS = 30
STATIC_PARAMS = {'boosting': 'gbdt',
'objective': 'binary',
'metric': 'auc',
}
N_TRIALS = 100
X = train_data.drop('Income', axis=1)
y = train_data.Income
def train_evaluate(X, y, params):
train_x, valid_x, train_y, valid_y = train_test_split( X, y, test_size=0.2, random_state=42)
train_data = lgb.Dataset(train_x, label = train_y)
valid_data = lgb.Dataset(valid_x, label= valid_y, reference=train_data)
model = lgb.train(params, train_data,
num_boost_round=NUM_BOOST_ROUND,
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
valid_sets=[valid_data],
valid_names=['valid'])
score = model.best_score['valid']['auc']
return score
def objective(trial):
params = {'learning_rate': trial.suggest_loguniform('learning_rate', 0.01, 0.5),
'max_depth': trial.suggest_int('max_depth', 1, 30),
'num_leaves': trial.suggest_int('num_leaves', 2, 100),
'min_data_in_leaf': trial.suggest_int('min_data_in_leaf', 10, 1000),
'feature_fraction': trial.suggest_uniform('feature_fraction', 0.1, 1.0),
'subsample': trial.suggest_uniform('subsample', 0.1, 1.0)}
all_params = {**params, **STATIC_PARAMS}
return train_evaluate(X, y, all_params)
#neptune.init(project='unltd148/bayeshyper')
neptune.init(project='unltd148/bayeshyper',
api_token='eyJhcGlfYWRkcmVzcyI6Imh0dHBzOi8vYXBwLm5lcHR1bmUuYWkiLCJhcGlfdXJsIjoiaHR0cHM6Ly9hcHAubmVwdHVuZS5haSIsImFwaV9rZXkiOiIxNzcwN2E0Yy05M2NkLTRjZTUtOGQ1MC01NTE2MGUyZjE2ODkifQ==') # your credentials
#neptune.create_experiment(name='bayes hpo')
#monitor = opt_utils.NeptuneMonitor()
#we want to maximize auc which is the value of the objective function
study = optuna.create_study(direction='maximize', study_name="BayeshpoExperiment", storage='sqlite:///bayeshpo.db')
study.optimize(objective, n_trials=N_TRIALS)
#study.optimize(objective, n_trials=N_TRIALS, callbacks=[monitor])
#opt_utils.log_study(study)
#neptune.stop()
https://app.neptune.ai/unltd148/bayeshyper/e/BAY-11
Remember to stop your run once you’ve finished logging your metadata (https://docs.neptune.ai/api-reference/run#stop). It will be stopped automatically only when the notebook kernel/interactive console is terminated.
[32m[I 2021-08-03 12:28:52,840][0m A new study created in RDB with name: BayeshpoExperiment[0m
[32m[I 2021-08-03 12:30:53,347][0m Trial 99 finished with value: 0.92880063092149 and parameters: {'learning_rate': 0.17700169345735636, 'max_depth': 4, 'num_leaves': 21, 'min_data_in_leaf': 63, 'feature_fraction': 0.4626178283293562, 'subsample': 0.5881713610049703}. Best is trial 95 with value: 0.9316501075106499.[0m
[85] valid's auc: 0.92828
[86] valid's auc: 0.928313
[87] valid's auc: 0.928509
[88] valid's auc: 0.928628
[89] valid's auc: 0.928731
[90] valid's auc: 0.928769
[91] valid's auc: 0.928757
[92] valid's auc: 0.928739
[93] valid's auc: 0.928746
[94] valid's auc: 0.928746
[95] valid's auc: 0.928746
[96] valid's auc: 0.928762
[97] valid's auc: 0.92873
[98] valid's auc: 0.92876
[99] valid's auc: 0.928773
[100] valid's auc: 0.928801
Did not meet early stopping. Best iteration is:
[100] valid's auc: 0.928801
trial = study.best_trial
print('AUC: {}'.format(trial.value))
print("Best hyperparameters: {}".format(trial.params))
AUC: 0.9316501075106499
Best hyperparameters: {'feature_fraction': 0.23353267069836048, 'learning_rate': 0.18207171684909748, 'max_depth': 8, 'min_data_in_leaf': 12, 'num_leaves': 37, 'subsample': 0.5551663236116008}
Below shows how to use Optuna library without the neptune.ai for hyperparameter optimization.
import lightgbm as lgb
import neptune.new as neptune
import neptune
import neptunecontrib.monitoring.optuna as opt_utils
import optuna
import pandas as pd
from sklearn.model_selection import train_test_split
import neptunecontrib.monitoring.optuna as opt_utils #
NUM_BOOST_ROUND = 100
EARLY_STOPPING_ROUNDS = 30
STATIC_PARAMS = {'boosting': 'gbdt',
'objective': 'binary',
'metric': 'auc',
}
N_TRIALS = 100
#data = pd.read_csv(TRAIN_PATH, nrows=N_ROWS)
X = train_data.drop('Income', axis=1)
y = train_data.Income
def train_evaluate(X, y, params):
train_x, valid_x, train_y, valid_y = train_test_split( X, y, test_size=0.2, random_state=42)
train_data = lgb.Dataset(train_x, label = train_y)
valid_data = lgb.Dataset(valid_x, label= valid_y, reference=train_data)
model = lgb.train(params, train_data,
num_boost_round=NUM_BOOST_ROUND,
early_stopping_rounds=EARLY_STOPPING_ROUNDS,
valid_sets=[valid_data],
valid_names=['valid'])
score = model.best_score['valid']['auc']
return score
def objective(trial):
params = {'learning_rate': trial.suggest_loguniform('learning_rate', 0.01, 0.5),
'max_depth': trial.suggest_int('max_depth', 1, 15),
'num_leaves': trial.suggest_int('num_leaves', 2, 100),
'min_data_in_leaf': trial.suggest_int('min_data_in_leaf', 10, 1000),
'n_estimators' : trial.suggest_int('n_estimators', 20, 100),
'feature_fraction': trial.suggest_uniform('feature_fraction', 0.1, 1.0),
'subsample': trial.suggest_uniform('subsample', 0.1, 1.0)}
all_params = {**params, **STATIC_PARAMS}
return train_evaluate(X, y, all_params)
study = optuna.create_study(direction='maximize', study_name="bayes_hpo_experiment", storage='sqlite:///bayeshpo.db')
study.optimize(objective, n_trials=N_TRIALS)
[67] valid's auc: 0.92876
Did not meet early stopping. Best iteration is:
[67] valid's auc: 0.92876
Trial 98 finished with value: 0.9287604448393214 and parameters: {'learning_rate': 0.11354649911022159, 'max_depth': 12, 'num_leaves': 91, 'min_data_in_leaf': 60, 'n_estimators': 67, 'feature_fraction': 0.49582158675330323, 'subsample': 0.2786627013714205}. Best is trial 95 with value: 0.9304994589047078.
Trial 98 finished with value: 0.9287604448393214 and parameters: {'learning_rate': 0.11354649911022159, 'max_depth': 12, 'num_leaves': 91, 'min_data_in_leaf': 60, 'n_estimators': 67, 'feature_fraction': 0.49582158675330323, 'subsample': 0.2786627013714205}. Best is trial 95 with value: 0.9304994589047078.
Trial 98 finished with value: 0.9287604448393214 and parameters: {'learning_rate': 0.11354649911022159, 'max_depth': 12, 'num_leaves': 91, 'min_data_in_leaf': 60, 'n_estimators': 67, 'feature_fraction': 0.49582158675330323, 'subsample': 0.2786627013714205}. Best is trial 95 with value: 0.9304994589047078.
[1] valid's auc: 0.895009
Training until validation scores don't improve for 30 rounds.
[2] valid's auc: 0.894377
[3] valid's auc: 0.90365
[4] valid's auc: 0.910059
[5] valid's auc: 0.91103
[6] valid's auc: 0.912663
[7] valid's auc: 0.913229
[8] valid's auc: 0.912465
[9] valid's auc: 0.914217
[10] valid's auc: 0.914101
[11] valid's auc: 0.91485
[12] valid's auc: 0.915569
[13] valid's auc: 0.916041
[14] valid's auc: 0.91614
[15] valid's auc: 0.916149
[16] valid's auc: 0.916778
[17] valid's auc: 0.916752
[18] valid's auc: 0.916654
[19] valid's auc: 0.917326
[20] valid's auc: 0.917528
[21] valid's auc: 0.917477
[22] valid's auc: 0.917588
[23] valid's auc: 0.917607
[24] valid's auc: 0.917942
[25] valid's auc: 0.91832
[26] valid's auc: 0.918393
[27] valid's auc: 0.918661
[28] valid's auc: 0.919599
[29] valid's auc: 0.920117
[30] valid's auc: 0.920364
[31] valid's auc: 0.920724
[32] valid's auc: 0.920832
[33] valid's auc: 0.920973
[34] valid's auc: 0.921119
[35] valid's auc: 0.921547
[36] valid's auc: 0.92176
[37] valid's auc: 0.922056
[38] valid's auc: 0.92246
[39] valid's auc: 0.922688
[40] valid's auc: 0.922936
[41] valid's auc: 0.922867
[42] valid's auc: 0.923032
[43] valid's auc: 0.923337
[44] valid's auc: 0.923505
[45] valid's auc: 0.923478
[46] valid's auc: 0.923805
[47] valid's auc: 0.923837
[48] valid's auc: 0.923978
[49] valid's auc: 0.924484
[50] valid's auc: 0.924439
[51] valid's auc: 0.924756
[52] valid's auc: 0.924842
[53] valid's auc: 0.925031
[54] valid's auc: 0.925212
[55] valid's auc: 0.925248
[56] valid's auc: 0.925575
[57] valid's auc: 0.925559
[58] valid's auc: 0.925678
[59] valid's auc: 0.925695
[60] valid's auc: 0.92588
[61] valid's auc: 0.926011
Did not meet early stopping. Best iteration is:
[61] valid's auc: 0.926011
[32m[I 2021-08-02 03:11:20,230][0m Trial 99 finished with value: 0.9260113046540378 and parameters: {'learning_rate': 0.072614609395996, 'max_depth': 11, 'num_leaves': 77, 'min_data_in_leaf': 99, 'n_estimators': 61, 'feature_fraction': 0.5745671761110418, 'subsample': 0.33137821810515283}. Best is trial 95 with value: 0.9304994589047078.[0m
Trial 99 finished with value: 0.9260113046540378 and parameters: {'learning_rate': 0.072614609395996, 'max_depth': 11, 'num_leaves': 77, 'min_data_in_leaf': 99, 'n_estimators': 61, 'feature_fraction': 0.5745671761110418, 'subsample': 0.33137821810515283}. Best is trial 95 with value: 0.9304994589047078.
Trial 99 finished with value: 0.9260113046540378 and parameters: {'learning_rate': 0.072614609395996, 'max_depth': 11, 'num_leaves': 77, 'min_data_in_leaf': 99, 'n_estimators': 61, 'feature_fraction': 0.5745671761110418, 'subsample': 0.33137821810515283}. Best is trial 95 with value: 0.9304994589047078.
Trial 99 finished with value: 0.9260113046540378 and parameters: {'learning_rate': 0.072614609395996, 'max_depth': 11, 'num_leaves': 77, 'min_data_in_leaf': 99, 'n_estimators': 61, 'feature_fraction': 0.5745671761110418, 'subsample': 0.33137821810515283}. Best is trial 95 with value: 0.9304994589047078.
The optimal values of the hyperparameters can be found once the tuning is completed.
trial = study.best_trial
print('AUC: {}'.format(trial.value))
print("Best hyperparameters: {}".format(trial.params))
AUC: 0.9304994589047078
Best hyperparameters: {'feature_fraction': 0.4670792062861959, 'learning_rate': 0.13696520601495382, 'max_depth': 12, 'min_data_in_leaf': 30, 'n_estimators': 68, 'num_leaves': 79, 'subsample': 0.2184961565013457}
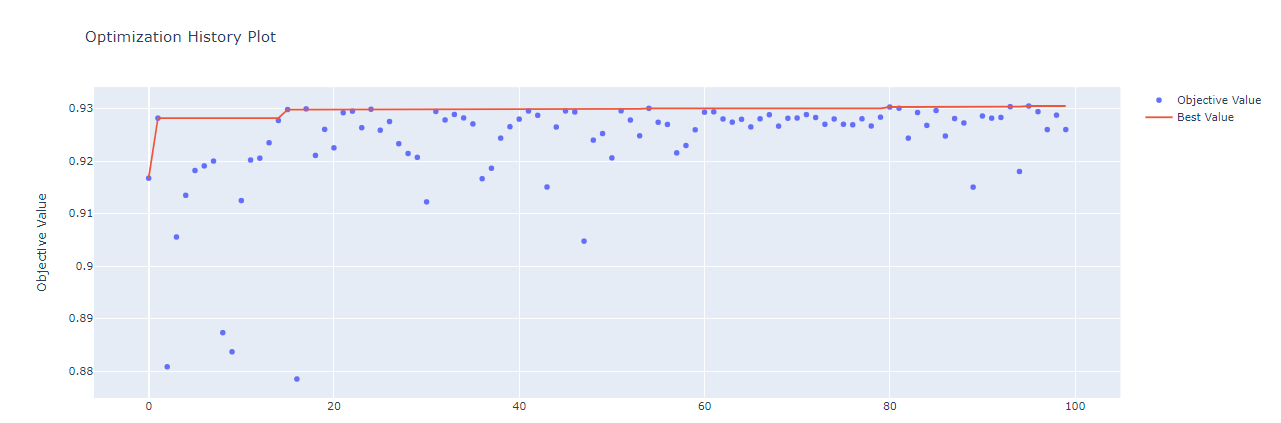
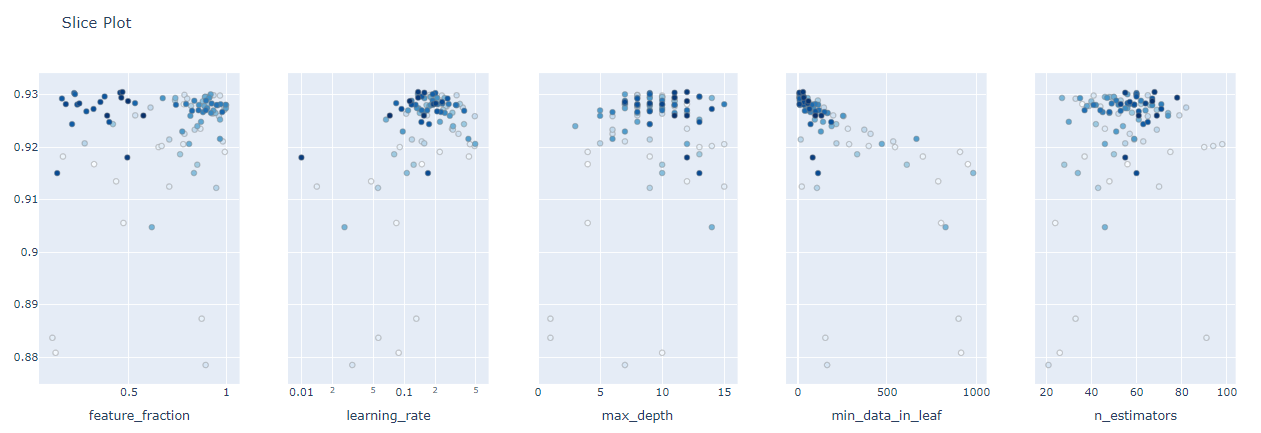
The highest AUC which is 0.93 together with the optimal values of the hyperparameters is shown above.There are options for different visualizations of the optimization process available in optuna. The one important for hyperparameter tuning are history plot, slice plot, contour plot, and parallel coordinate plot.
optuna.visualization.plot_optimization_history(study)
optuna.visualization.plot_slice(study)
4. Hyperopt
Hyperopt is an open source hyperparameter optimization library that implements bayesian optimization for hyperparameter tuning as well as other algorithms. Hyperopt implements three algorithms Random Search, Random Search, Tree of Parzen Estimators (TPE) and Adaptive TPE. The bayesian optimizer is implemented as TPE. More information on how to get started with this library is found here. There is also the Hyperopt-sklearn which is Hyperopt-based model selection among machine learning algorithms wrapper for scikit-learn.
# Run an XGBoost model with hyperparmaters that are optimized using hyperopt
# The output of the script are the best hyperparmaters
# The optimization part using hyperopt is partly inspired from the following script:
# https://github.com/bamine/Kaggle-stuff/blob/master/otto/hyperopt_xgboost.py
import pandas as pd
import numpy as np
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
from hyperopt import STATUS_OK, Trials, fmin, hp, tpe
#-------------------------------------------------#
X = train_data.drop('Income', axis=1)
y = train_data.Income
train_x, valid_x, train_y, valid_y = train_test_split( X, y, test_size=0.2, random_state=42)
print('The training set is of length: ', len(train_y))
print('The validation set is of length: ', len(valid_y))
#-------------------------------------------------#
# Utility functions
def intersect(l_1, l_2):
return list(set(l_1) & set(l_2))
def get_features(train, test):
intersecting_features = intersect(train.columns, test.columns)
intersecting_features.remove('people_id')
intersecting_features.remove('activity_id')
return sorted(intersecting_features)
#-------------------------------------------------#
# Scoring and optimization functions
def score(params):
print("Training with params: ")
print(params)
num_round = int(params['n_estimators'])
del params['n_estimators']
dtrain = xgb.DMatrix(train_x, label=train_y)
dvalid = xgb.DMatrix(valid_x, label=valid_y)
watchlist = [(dvalid, 'eval'), (dtrain, 'train')]
gbm_model = xgb.train(params, dtrain, num_round,
evals=watchlist,
verbose_eval=True)
predictions = gbm_model.predict(dvalid,
ntree_limit=gbm_model.best_iteration + 1)
score = roc_auc_score(valid_y, predictions)
# TODO: Add the importance for the selected features
print("\tScore {0}\n\n".format(score))
# The score function should return the loss (1-score)
# since the optimize function looks for the minimum
loss = 1 - score
return {'loss': loss, 'status': STATUS_OK}
def optimize(
#trials,
random_state=SEED):
"""
This is the optimization function that given a space (space here) of
hyperparameters and a scoring function (score here), finds the best hyperparameters.
"""
# To learn more about XGBoost parameters, head to this page:
# https://github.com/dmlc/xgboost/blob/master/doc/parameter.md
space = {
'n_estimators': hp.quniform('n_estimators', 100, 1000, 1),
'eta': hp.quniform('eta', 0.025, 0.5, 0.025),
# A problem with max_depth casted to float instead of int with
# the hp.quniform method.
'max_depth': hp.choice('max_depth', np.arange(1, 14, dtype=int)),
'min_child_weight': hp.quniform('min_child_weight', 1, 6, 1),
'subsample': hp.quniform('subsample', 0.5, 1, 0.05),
'gamma': hp.quniform('gamma', 0.5, 1, 0.05),
'colsample_bytree': hp.quniform('colsample_bytree', 0.5, 1, 0.05),
'eval_metric': 'auc',
'objective': 'binary:logistic',
# Increase this number if you have more cores. Otherwise, remove it and it will default
# to the maxium number.
'nthread': 4,
'booster': 'gbtree',
'tree_method': 'exact',
'silent': 1,
'seed': random_state
}
# Use the fmin function from Hyperopt to find the best hyperparameters
best = fmin(score, space, algo=tpe.suggest,
# trials=trials,
max_evals=250)
return best
#-------------------------------------------------#
#-------------------------------------------------#
# Run the optimization
# Trials object where the history of search will be stored
# For the time being, there is a bug with the following version of hyperopt.
# You can read the error messag on the log file.
# For the curious, you can read more about it here: https://github.com/hyperopt/hyperopt/issues/234
# => So I am commenting it.
# trials = Trials()
best_hyperparams = optimize(
#trials
)
print("The best hyperparameters are: ", "\n")
print(best_hyperparams)
[736] eval-auc:0.926821 train-auc:0.951919
[737] eval-auc:0.926821 train-auc:0.951919
[738] eval-auc:0.926821 train-auc:0.951919
[739] eval-auc:0.926821 train-auc:0.951919
[740] eval-auc:0.926821 train-auc:0.951919
[741] eval-auc:0.926821 train-auc:0.951919
[742] eval-auc:0.926821 train-auc:0.951919
[743] eval-auc:0.926821 train-auc:0.951919
[744] eval-auc:0.926821 train-auc:0.951919
Score 0.9268210155692731
Training with params:
{'booster': 'gbtree', 'colsample_bytree': 0.75, 'eta': 0.225, 'eval_metric': 'auc', 'gamma': 0.6000000000000001, 'max_depth': 9, 'min_child_weight': 2.0, 'n_estimators': 916.0, 'nthread': 4, 'objective': 'binary:logistic', 'seed': 314159265, 'silent': 1, 'subsample': 0.9500000000000001, 'tree_method': 'exact'}
[0] eval-auc:0.907075 train-auc:0.908109
[1] eval-auc:0.910798 train-auc:0.917816
81%|████████ | 202/250 [2:12:13<40:56, 51.17s/it, best loss: 0.06961736409698138]
"best hyperparameters : {}".format(best_hyperparams)
best hyperparameters : {‘colsample_bytree’: 0.55, ‘eta’: 0.025, ‘gamma’: 0.55, ‘max_depth’: 6, ‘min_child_weight’: 2.0, ‘n_estimators’: 540.0, ‘subsample’: 0.8}
References
[1] Turner et al Bayesian Optimization is Superior to Random Search for Machine Learning Hyperparameter Tuning: Analysis of the Black-Box Optimization Challenge 2020 [2] https://scikit-optimize.github.io/stable/ [3] https://optuna.org/ [4] http://hyperopt.github.io/hyperopt/